The accurate replication of genetic material DNA is one of the most fundamental biological processes essential for the survival, growth, and reproduction of all living organisms. Eukaryotes, including humans, plants, yeast, and paramecia, share a similarly complex DNA replication machinery. How did this sophisticated machinery evolve? A team led by Associate Professor Fabai Wu from the College of Science at Eastern Institute of Technology, Ningbo has found answers in a group of marine archaea.
On October 21st (Beijing Time), the related research findings were published in Nature Ecology & Evolution. Eastern Institute of Technology, Ningbo is the first completing institution.
All cellular life, from simple prokaryotes to more complex eukaryotes, uses double-stranded DNA as its genetic material. DNA replication errors serve as both a driving force for evolution and a major contributor to diseases like cancer.
Complex eukaryotes evolved from simpler prokaryotes (archaea). However, eukaryotic genomes are typically tens to thousands of times larger than those of prokaryotes. To ensure the accurate replication of these vast genomes, eukaryotes possess a DNA replication machinery that is far more complex than that of their ancestors, featuring mechanisms such as the specialized division of labor among multiple DNA polymerases. This complexity has long been considered unique to early eukaryotes.
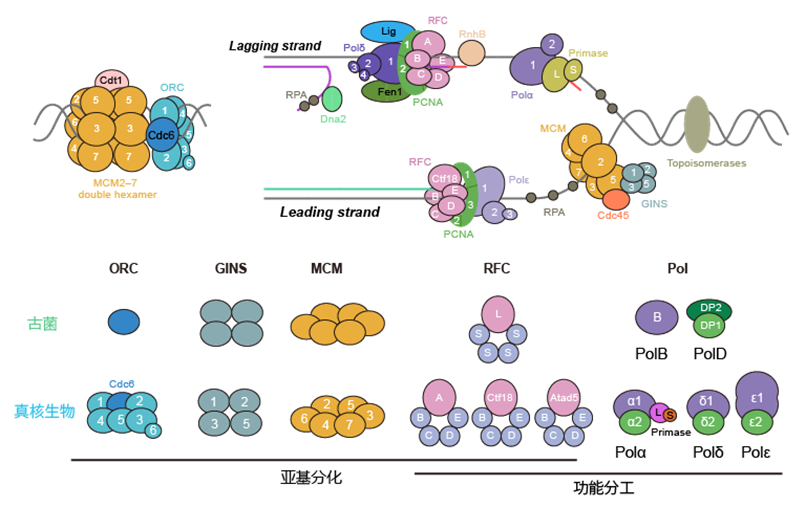
Eukaryotic DNA replication components evolved from the complexification of archaeal components | Image provided by the research team
Professor Fabai Wu's team discovered that multiple complex modules were already present in prokaryotes of the Asgard archaea. Consequently, they propose that the complexification of the core genetic machinery in the Last Eukaryotic Common Ancestor (LECA) began within the archaeal lineage. These findings fill a long-standing knowledge gap in the field of genetics and also provide new perspectives for the future biotechnological development of archaeal DNA polymerases.
While the scientific community previously widely believed this complexity was unique to eukaryotes, this research, through systematic analysis of extensive genomic data from archaea and eukaryotes, demonstrates that the functional specialization and structural complexification of some replication machinery components originated in Asgard archaea, predating the origin of eukaryotes.
Additionally, the study indicates that Horizontal Gene Transfer (HGT) was a key driver of core genetic innovation during eukaryotic origins. Although HGT is recognized as an important pathway for acquiring new functions, its role in shaping core genetic mechanisms has not received sufficient attention. The researchers propose that the infiltration of core functional genes via HGT played a crucial role in the complexification of genetic mechanisms during the evolution from archaea to eukaryotes.
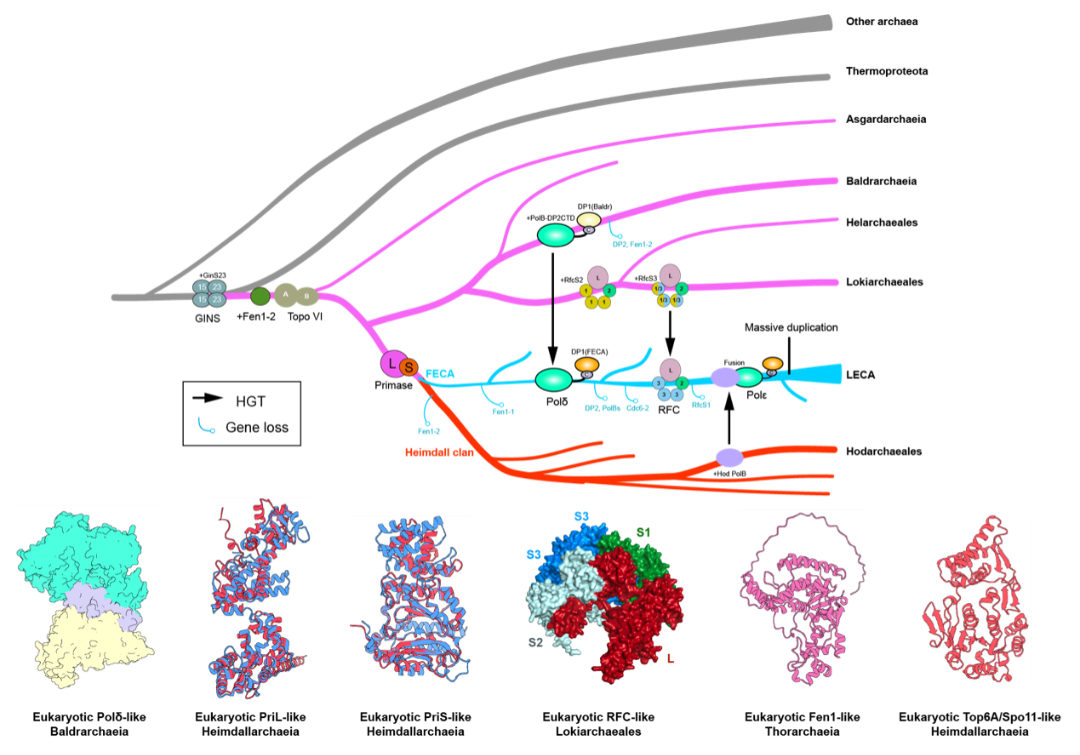
Evolutionary trajectory of DNA replication machinery during eukaryotic origins | Image provided by the research team
The first author of the paper is Associate Researcher Yanlei Feng from the ZJU-Hangzhou Global Scientific and Technological Innovation Center. The corresponding author is Associate Professor Fabai Wu from Eastern Institute of Technology. Other contributing team members include Associate Researcher Jingjing Ding from Hangzhou Polytechnic University, graduate student Youxiong Lin, and research assistant Danxi Cui from the ZJU-Hangzhou Global Scientific and Technological Innovation Center. Collaborators on this work include Professor Daoqiong Zheng and graduate student Kejing Li from the Ocean College of Zhejiang University, Chair Professor Zongwei Cai from Eastern Institute of Technology, Ningbo, and Professor Stephen D. Bell from The Ohio State University, USA. This research received support from the National Natural Science Foundation of China (NSFC), the Ningbo Key Laboratory of Biomedical Data Mining and Computation, the Ministry of Science and Technology's ONCE program, as well as other funding sources.
Link: https://doi.org/10.1038/s41559-025-02882-6